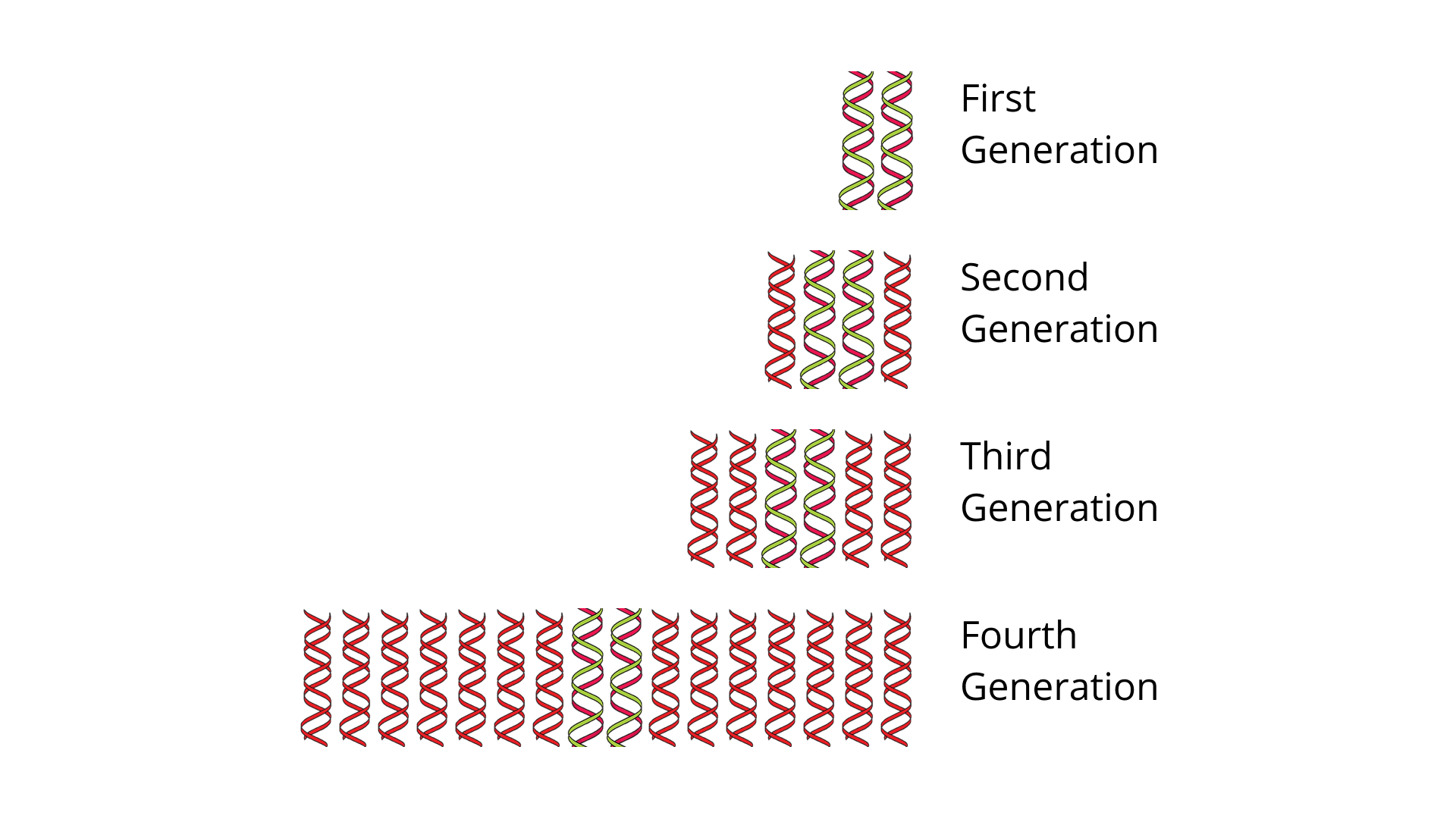
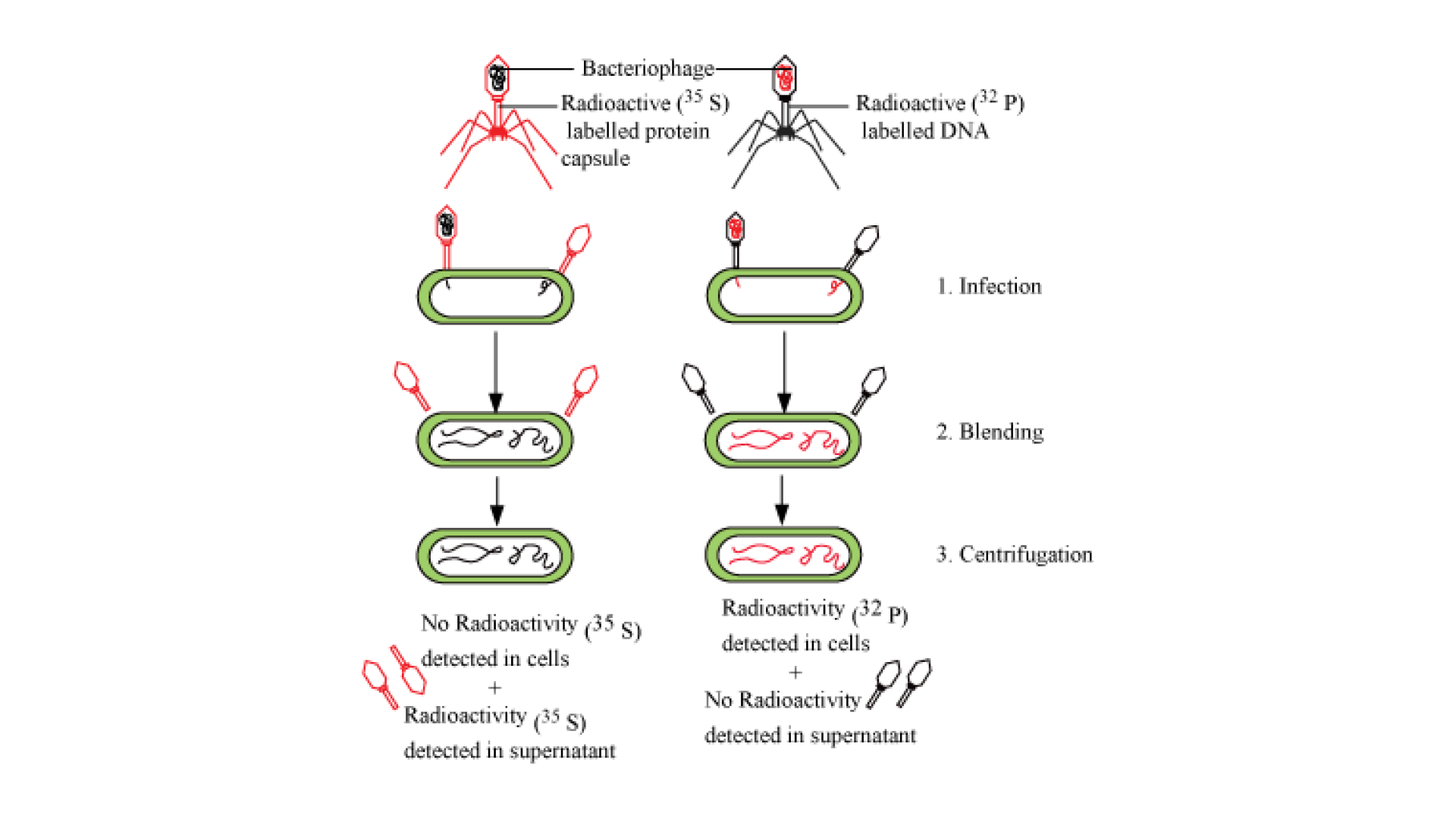
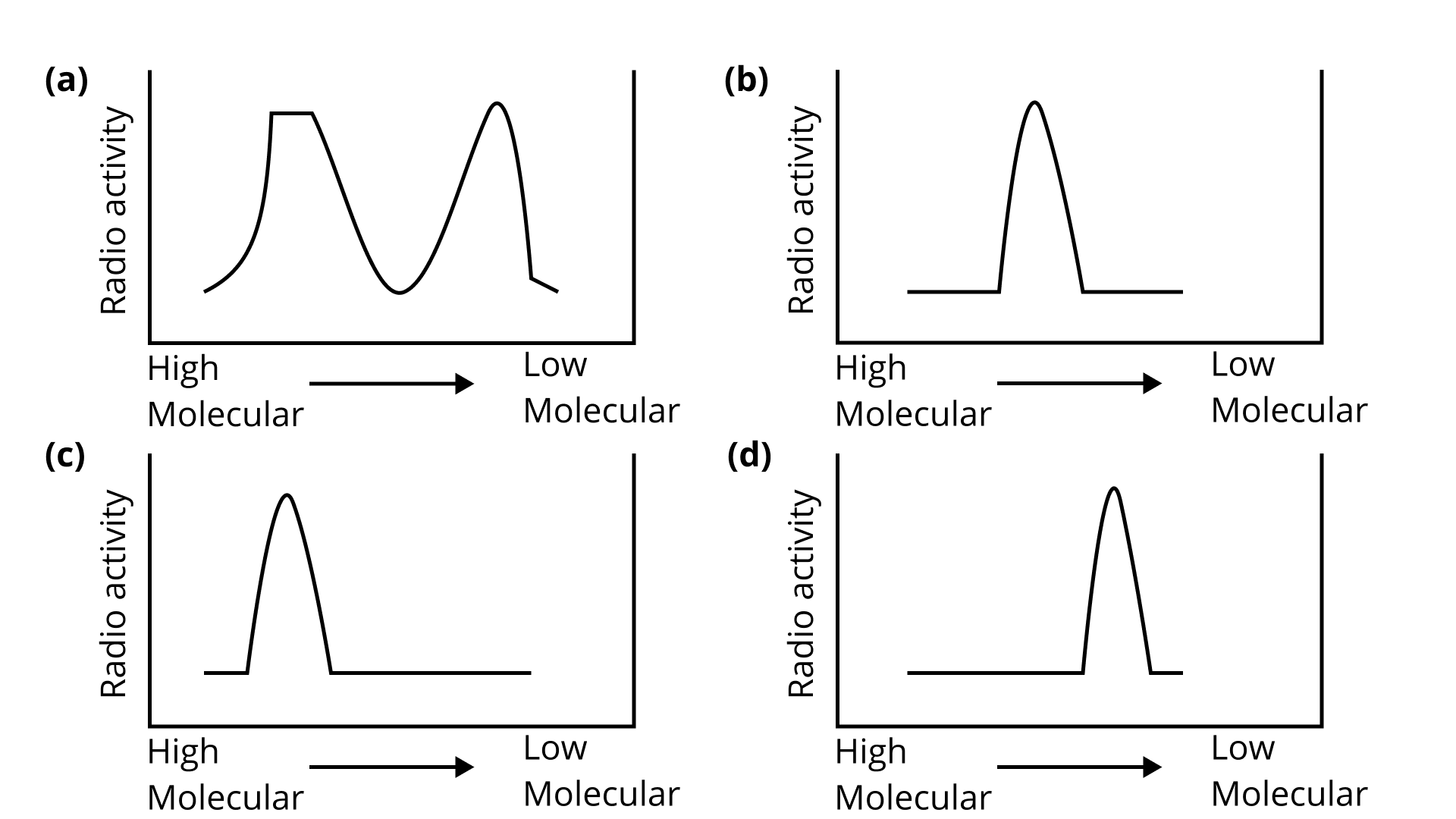
Download Free PDF of NCERT Exemplar for Class 12 Biology - Molecular Basis of Inheritance
FAQs on NCERT Exemplar for Class 12 Biology Chapter-6 (Book Solutions)
1. What is an account of post-transcriptional modifications of a eukaryotic mRNA given in the NCERT book?
Post-transcriptional modifications are the chemical changes accomplished to the number one transcript of RNA. These adjustments convert a gene into practical RNA.
In prokaryotes, the number one transcript (hn-RNA) has each the non-coding introns inside the gene and the coding exons but they are non-practical. Hence, Splicing takes region. In a very correct order introns are removed and exons are connected. In Eukaryotes (in-RNA), which is also known as the primary transcript is not present so the reduction isn't needed. In this capping of Methyl Guanosine Triphosphate nucleotide is introduced to the five’ quit of hn-RNA called cap shape and in tailing addition of Adenylate residues at the 3’ end of the RNA takes the vicinity. Hn-RNA is processed to shape mRNA or messenger RNA after all this technique and accordingly transported out of the nucleus for translation.
2. What is operon? Giving an example, explain an Inducible operon concerning Chapter 6 NCERT biology.
The operon is a collection of a gene that has associated features and is inside the catabolism or degradation of lactose which turned into given with the aid of François Jacob and Jacques Monad in 1961.
Inducer operon also referred to as Lac Operon (lactose) and Trp Operon additionally called Repressor operon (Tryptophan operon) are the two forms of the operon.
Inducible operon system:
Lac Operon is determined in bacteria E. Coli. When E. Coli infests on something it prefers Glucose over lactose.
Glucose is taken up by using E. Coli, and it starts the use of Lactose.
When the consumption of lactose is commenced employing E. Coli, Lac operon gets activated which proves that lactose is an inducer for the Lac Operon.
Lac Operon
Gene ‘i’ undergo transcription In the operon and paperwork a messenger RNA or mRNA. This undergoes translation to form repressor protein.
A repressor tetramer is fashioned employing combining the repressor protein and the promoter web page or gene ‘a is the binding web page for enzyme RNA polymerase.
As the repressor tetramer movements toward the operator web page to bind to it, lactose present inside the E. Coli breaks down the tetramer. The operator receives unblocked and makes it in a position for enzyme RNA polymerase to transport forward from the promoter website.
This allows the enzyme RNA polymerase to begin transcribing structural genes z,y, and a. Gene ‘z’ coding for enzyme b-galactosidase break down into glucose and lactose. Gene ‘y’ coding for enzyme permease offers the entry of more lactose in the bacteria E.Coli.
3. What are the six features of the human genome given in Chapter 6 Molecular Basis of inheritance?
The genome of the Homo Sapiens. Human Genome. It consists of the coding areas of the DNA which encodes all of the genes. It also consists of all the non-coding areas of the DNA which does not encode for any genes.
Six Capabilities of the Human Genome are:
The genome has around 3164.7 million nucleotide bases and
99.9 % of the nucleotide bases are the same in all humans.
The largest gene is Dystrophin having 2.4 million bases.
Chromosome 1 has the most genes (2968) and Y has the least genes (231).
Less than 2% of the genome has the coding series for proteins.
Only 50% of the whole discovered genes have recognized functions.
4. What are the methods used in sequencing the human genome according to NCERT Exemplar for Class 12?
DNA sequencing is the approach used to discover the definite order of bases (Adenosine, Guanine, Thymine, and Cytosine) in the DNA.
A) Maxam Gilbert Method
B) Sanger Method
These are the 2 fundamental strategies.
Maxam Gilbert Method: In 1977 Maxim Gilbert devised for DNA sequencing
1. By growing the temperature the DNA denatured into unmarried-stranded DNA. The five’ stop is categorized radioactively via Kinase response using gamma P 32. Cleave the DNA strand at unique positions the use of chemical reactions. For cleavage at unique positions, chemical compounds are used observed by the addition of piperidine. Dimethyl Sulphate: This chemical simplest attacks purines (Adenosine and Guanine). Hydrazine: This chemical most effective attacks pyrimidine (Cytosine and Thymine) In Maxim and Gilbert test the chemicals cleaved G, A+G, C, and C+T. In four take a look at tubes, four exclusive sizes of DNA are acquired. Now, those DNA’s are separated based on length by way of gel electrophoresis approach. After this loss of connection, the sequence of DNA is received.
5. What is the process of translation and where can I find the explanation on Vedantu?
The translation is the interpreting of mRNA to form an amino acid chain with the help of ribosomes within the cytosol of the cell.
Amino acids, m-RNA, t-RNA, and Ribosomes are worried in this manner.
Initiation:
1. At the five’ stop of the mRNA, the smaller 40S subunit of the ribosome with methionyl-tRNA scans the mRNA to find the begin codon (5’AUG) and particular to methionine.
2. The larger 60S subunit of the ribosome binds to the mRNA.This 60S ribosomal subunit has t-RNA binding sites.
Site P: This site can keep the peptide chain.
Site A: This web page can hold t-RNA.
Elongation:
1. As the Met-tRNA binds to the P site of the 60S subunit of the RNA. Another aminoacyl- tRNA and it is complementary to the next codon occupies website A inside the 60S subunit of the ribosome.
2. In the middle of Methionyl-tRNA and Aminoacyl-tRNA peptidyl transferase paperwork a peptide bond.
3. T-RNA molecule on the P site turn out to be uncharged and leaves the ribosome. Hence, it moves along the mRNA molecule to the next codon which opens Site A for the following aminoacyl-tRNA.
4. The polypeptide chain is constructed inside the direction of the N to the C terminal.
Termination:
The forestall codon enters the site because the A site is vacant. None of the t-RNA molecules binds to this codon.
The peptide bond of Methionyl-tRNA and Aminoacyl-tRNA end up hydrolyzed releasing the polypeptide into the cytoplasm. The dissociation of the ribosomal subunit takes region.
You can also find these solutions in the Vedantu app where professional teachers give LIVE lectures for you to understand better and score well in both your 12th and NEET.