
Write an essay on recombinant DNA technology.
Answer
565.2k+ views
Hint:Recombinant DNA (rDNA) is a DNA molecule that is cut and inserted with the desired gene or protein. This rDNA is then amplified to get a large quantity of that desired gene fragment or product encoded by that gene.
Complete answer: Deoxyribonucleic acid (DNA) is a molecule that carries the genetic information of a particular organism. A DNA molecule has a double helix structure has two antiparallel strands made up of deoxyribose sugar and a phosphate group. Each sugar is one of the four bases adenine-A, cytosine-C, guanine-G, and thymine-T where A pairs with T and G pairs with C. This sequence contains information for protein and gene expression.
Recombinant DNA technology is a branch of genetic engineering where DNA molecules of an organism are altered by inserting new fragments of DNA (gene or protein) from other organisms to get a recombinant DNA (rDNA) with desired traits. This technique involves identification, isolation, and insertion of desired DNA fragment or gene of interest into a vector such as a plasmid or bacteriophage to form an rDNA molecule that is further multiplied to produce a large amount of desired gene or its product.
Steps involved in rDNA technology:
Step 1:Identifying and isolating the desired gene of interest
In rDNA technology, there are different resources to get the desired gene of interest of fragment of DNA that include Genomic library, cDNA library, if we know the sequence of the desired gene then we can chemically synthesize that gene n use it for rDNA technology, also if the number of copies desired gene are not enough we can use PCR (Polymerase chain reaction) to amplify them. Let’s understand rDNA technology with an example:
We will now consider a human insulin gene which is to be introduced into E.coli host cells through a vector plasmid.
So in the first step, we will get the human insulin gene from the genomic library.
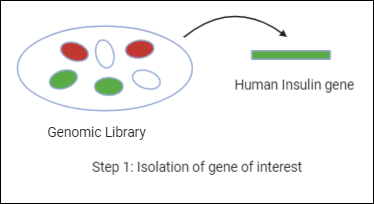
Step 2: Inserting the isolated gene into a suitable vector
A vector is any DNA molecule that has the ability to replicate inside the host to which the desired gene has integrated for cloning. Vectors include plasmids, bacteriophages, cosmids, BAC (Bacterial artificial chromosome), yeast vectors, etc.
Let’s consider plasmid as a vector for our example, so now we will isolate plasmid that is an extrachromosomal DNA present in E.coli cells. This isolated plasmid is then subjected to restriction enzymes (Enzymes that are capable of making cuts at specific sites in the DNA molecule) that cut the DNA molecule and now we can introduce our insulin gene in the vector and we can seal it with the help of enzyme ligase. Now, we can call this DNA molecule a recombinant DNA molecule or chimeric DNA.
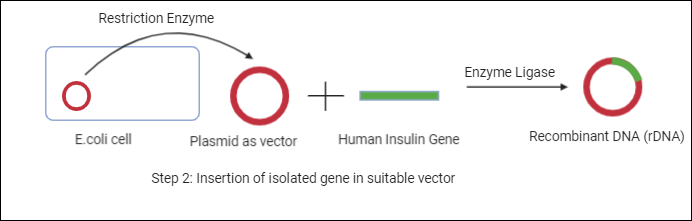
Step 3: Introduction of this vector into a suitable host
There are many methods to introduce our vector into the host cells:
Physical gene transfer methods like electroporation, Microinjection, DNA transfer via pollen, etc.
Chemical gene transfer methods like Polyethylene glycol mediated (PEG mediated), Calcium chloride mediated, DEAE dextran mediated gene transfer, or certain cells like bacterial cells are capable of taking genes from the surrounding this is called Transformation.
Coming back to our example, we have an rDNA molecule that is needed to introduce in a host cell. The host cell will be mostly a bacterial cell (i.e. E.coli cell) as it is very easy to manipulate. Now, we have a genetically modified E.coli cell with a human insulin gene.
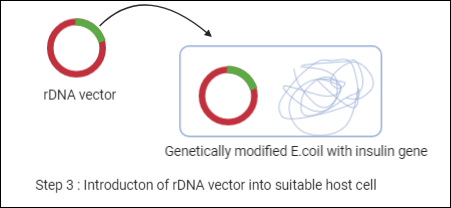
Step 4: Selection of the transformed host cell
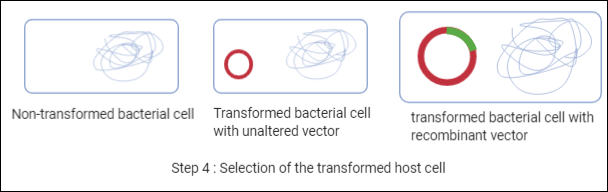
After step 3, we will have three types of cell colonies one with non-transformed bacterial cell here there will be no change in E.coli cell, second will be transformed bacterial cell with unaltered vector where the cell has transformed but without our gene of interest, and the third will be transformed bacterial cell with recombinant vector. Colonies of the third type will be selected. Selection can be done by growing antibiotic resistance in a selective medium, or if the desired gene shows any visible characters based on that, or if it is an enzyme we can use assay, etc.
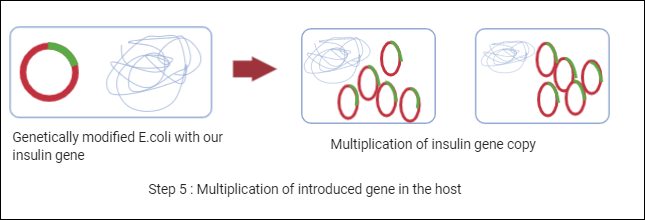
Step 5: Multiplication of introduced desired gene in the host
Now, we have an E.coli cell with a human insulin gene. These plasmids will replicate inside the host and the bacteria will also replicate making millions of copies of this insulin gene. This gene will further undergo transcription and translation to synthesize insulin.
In this way we can produce large amounts of insulin in bacterial cells, this is called recombinant DNA technology.
Note: While inserting desired genes in vectors, restriction enzymes that are used have specific restriction sites that cut the molecules at that site. Hence, it is important to locate the site where the gene is to be inserted and accordingly restriction enzymes are to be selected. There are few restriction enzymes that can cut the molecule at more than two sites. An example of restriction enzymes is EcoRI.
Complete answer: Deoxyribonucleic acid (DNA) is a molecule that carries the genetic information of a particular organism. A DNA molecule has a double helix structure has two antiparallel strands made up of deoxyribose sugar and a phosphate group. Each sugar is one of the four bases adenine-A, cytosine-C, guanine-G, and thymine-T where A pairs with T and G pairs with C. This sequence contains information for protein and gene expression.
Recombinant DNA technology is a branch of genetic engineering where DNA molecules of an organism are altered by inserting new fragments of DNA (gene or protein) from other organisms to get a recombinant DNA (rDNA) with desired traits. This technique involves identification, isolation, and insertion of desired DNA fragment or gene of interest into a vector such as a plasmid or bacteriophage to form an rDNA molecule that is further multiplied to produce a large amount of desired gene or its product.
Steps involved in rDNA technology:
Step 1:Identifying and isolating the desired gene of interest
In rDNA technology, there are different resources to get the desired gene of interest of fragment of DNA that include Genomic library, cDNA library, if we know the sequence of the desired gene then we can chemically synthesize that gene n use it for rDNA technology, also if the number of copies desired gene are not enough we can use PCR (Polymerase chain reaction) to amplify them. Let’s understand rDNA technology with an example:
We will now consider a human insulin gene which is to be introduced into E.coli host cells through a vector plasmid.
So in the first step, we will get the human insulin gene from the genomic library.

Step 2: Inserting the isolated gene into a suitable vector
A vector is any DNA molecule that has the ability to replicate inside the host to which the desired gene has integrated for cloning. Vectors include plasmids, bacteriophages, cosmids, BAC (Bacterial artificial chromosome), yeast vectors, etc.
Let’s consider plasmid as a vector for our example, so now we will isolate plasmid that is an extrachromosomal DNA present in E.coli cells. This isolated plasmid is then subjected to restriction enzymes (Enzymes that are capable of making cuts at specific sites in the DNA molecule) that cut the DNA molecule and now we can introduce our insulin gene in the vector and we can seal it with the help of enzyme ligase. Now, we can call this DNA molecule a recombinant DNA molecule or chimeric DNA.

Step 3: Introduction of this vector into a suitable host
There are many methods to introduce our vector into the host cells:
Physical gene transfer methods like electroporation, Microinjection, DNA transfer via pollen, etc.
Chemical gene transfer methods like Polyethylene glycol mediated (PEG mediated), Calcium chloride mediated, DEAE dextran mediated gene transfer, or certain cells like bacterial cells are capable of taking genes from the surrounding this is called Transformation.
Coming back to our example, we have an rDNA molecule that is needed to introduce in a host cell. The host cell will be mostly a bacterial cell (i.e. E.coli cell) as it is very easy to manipulate. Now, we have a genetically modified E.coli cell with a human insulin gene.

Step 4: Selection of the transformed host cell
After step 3, we will have three types of cell colonies one with non-transformed bacterial cell here there will be no change in E.coli cell, second will be transformed bacterial cell with unaltered vector where the cell has transformed but without our gene of interest, and the third will be transformed bacterial cell with recombinant vector. Colonies of the third type will be selected. Selection can be done by growing antibiotic resistance in a selective medium, or if the desired gene shows any visible characters based on that, or if it is an enzyme we can use assay, etc.

Step 5: Multiplication of introduced desired gene in the host
Now, we have an E.coli cell with a human insulin gene. These plasmids will replicate inside the host and the bacteria will also replicate making millions of copies of this insulin gene. This gene will further undergo transcription and translation to synthesize insulin.

In this way we can produce large amounts of insulin in bacterial cells, this is called recombinant DNA technology.
Note: While inserting desired genes in vectors, restriction enzymes that are used have specific restriction sites that cut the molecules at that site. Hence, it is important to locate the site where the gene is to be inserted and accordingly restriction enzymes are to be selected. There are few restriction enzymes that can cut the molecule at more than two sites. An example of restriction enzymes is EcoRI.
Recently Updated Pages
Master Class 12 Economics: Engaging Questions & Answers for Success

Master Class 12 Physics: Engaging Questions & Answers for Success

Master Class 12 English: Engaging Questions & Answers for Success

Master Class 12 Social Science: Engaging Questions & Answers for Success

Master Class 12 Maths: Engaging Questions & Answers for Success

Master Class 12 Business Studies: Engaging Questions & Answers for Success

Trending doubts
Which are the Top 10 Largest Countries of the World?

What are the major means of transport Explain each class 12 social science CBSE

Draw a labelled sketch of the human eye class 12 physics CBSE

Why cannot DNA pass through cell membranes class 12 biology CBSE

Differentiate between insitu conservation and exsitu class 12 biology CBSE

Draw a neat and well labeled diagram of TS of ovary class 12 biology CBSE




