
Step-by-Step Formation and Joining of Okazaki Fragments in DNA Replication
The concept of Okazaki fragments is essential in biology and helps explain real-world biological processes and exam-level questions effectively, especially for NEET. Understanding Okazaki fragments improves clarity on how DNA replication occurs and prevents common mistakes during exam preparation.
Understanding Okazaki Fragments
Okazaki fragments refer to the short DNA segments that are synthesized discontinuously on the lagging strand during DNA replication. This concept is important in areas like DNA replication, cell division, and molecular genetics. Okazaki fragments help ensure that both strands of DNA are accurately copied, even though DNA polymerase can only add nucleotides in one direction (5’ to 3’).
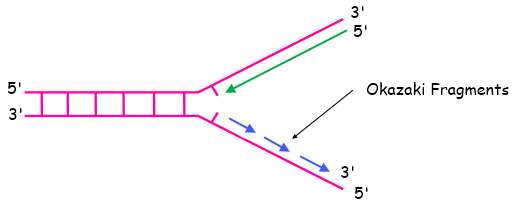
Mechanism of Okazaki Fragments
The basic mechanism involves the following steps:
- When DNA unwinds at the replication fork, one strand (leading strand) is made continuously in the 5’ to 3’ direction.
- The lagging strand runs in the opposite direction (3’ to 5’), so it cannot be synthesized continuously.
- Instead, the lagging strand is made in short, discontinuous segments called Okazaki fragments.
- Each Okazaki fragment starts with a short RNA primer laid down by primase.
- DNA polymerase extends the fragment in the 5’ to 3’ direction.
- After fragments are synthesized, DNA polymerase I removes the RNA primers and fills the gaps with DNA.
- Finally, DNA ligase joins the Okazaki fragments into a continuous strand.
Here’s a helpful table to understand Okazaki fragments better:
Okazaki Fragments Table
| Aspect | Description | Enzyme Involved |
|---|---|---|
| Location | Lagging strand of DNA during replication | DNA polymerase, primase, ligase |
| Length | Several hundred to a few thousand nucleotides | - |
| Initiation | By RNA primer | Primase |
| Joining | Fragments joined to form continuous strand | DNA ligase |
Worked Example – Biological Process
Let’s understand the process step by step:
1. As the DNA double helix unwinds at the replication fork, the leading strand is synthesized continuously toward the fork.
2. The lagging strand template runs in a 5' to 3' direction away from the fork, making continuous synthesis impossible for DNA polymerase.
3. Primase adds an RNA primer, and DNA polymerase extends each fragment from the primer, producing an Okazaki fragment.
4. When enough DNA has unwound, another fragment is started with a new primer.
5. DNA polymerase I removes each RNA primer and replaces it with DNA.
6. DNA ligase then joins all the fragments to produce a single, continuous DNA molecule.
Final Understanding: Okazaki fragments solve the challenge of antiparallel DNA strand orientation during replication.
Practice Questions
- What is the function of Okazaki fragments during DNA replication?
- Name the enzyme that joins Okazaki fragments on the lagging strand.
- Why are Okazaki fragments needed on the lagging strand but not on the leading strand?
- Draw and label a diagram showing the formation of Okazaki fragments.
- How does the length of Okazaki fragments differ in prokaryotes and eukaryotes?
Common Mistakes to Avoid
- Confusing Okazaki fragments with RNA primers.
- Writing that Okazaki fragments form on the leading strand (they only form on the lagging strand).
- Getting the direction of DNA synthesis (5’ to 3’) incorrect for Okazaki fragments.
- Forgetting the role of DNA ligase in joining the fragments.
Real-World Applications
The concept of Okazaki fragments is used in fields like medicine, biotechnology, and genetic engineering. For example, errors in fragment processing can lead to mutations. Techniques in molecular biology, such as DNA sequencing and polymerase chain reaction, also rely on the principles of fragment synthesis. Vedantu helps students relate such topics to practical examples and NEET MCQs.
In this article, we explored Okazaki fragments, their formation, relevance to DNA replication, related enzymes, and real-life significance. Keep practicing related NEET questions to build strong foundational concepts and accuracy. For more on DNA replication and genetics, review the related Vedantu resources below.
- DNA Replication
- Difference Between Replication and Transcription
- Cell Structure and Function
- Molecular Basis of Inheritance
- DNA Structure
- Neet Biology MCQs
- Difference Between Prokaryotic and Eukaryotic DNA Replication
- Difference Between Introns and Exons
- Principles of Inheritance and Variation
FAQs on What are Okazaki Fragments?
1. What is an Okazaki fragment in NEET?
An Okazaki fragment is a short segment of DNA that is synthesized discontinuously on the lagging strand during DNA replication. These fragments enable DNA polymerase to replicate the strand in the correct 5' to 3' direction despite the overall strand orientation being 3' to 5'.
2. Why are Okazaki fragments formed on the lagging strand?
Okazaki fragments form on the lagging strand because DNA polymerase can only synthesize DNA in the 5' to 3' direction, but the lagging strand template is oriented 5' to 3'. Thus, synthesis happens in short discontinuous segments opposite to the replication fork movement.
3. Which enzymes connect Okazaki fragments?
The enzyme DNA ligase joins Okazaki fragments by sealing the sugar-phosphate backbone, creating a continuous DNA strand. Before that, DNA polymerase I removes RNA primers and fills in the gaps with DNA.
4. How do Okazaki fragments differ in prokaryotes and eukaryotes?
While the fundamental process of Okazaki fragment synthesis is conserved, in prokaryotes Okazaki fragments are generally shorter (about 1000–2000 nucleotides), and in eukaryotes, they are shorter still (about 100–200 nucleotides). The enzymes involved may also differ slightly in complexity and regulation due to cellular differences.
5. How are Okazaki fragments important for DNA replication?
Okazaki fragments are crucial as they enable replication of the antiparallel lagging strand during DNA duplication. Without these fragments, DNA polymerase could not synthesize the lagging strand, preventing accurate and complete replication essential for cell division and genetic inheritance.
6. Why do students confuse which strand Okazaki fragments form on?
Confusion arises because of the antiparallel nature of DNA strands and the opposing directions of replication. Remember that Okazaki fragments always form on the lagging strand, which is synthesized discontinuously opposite to the replication fork’s movement, unlike the continuous leading strand. Visualizing strand directionality helps reduce this error.
7. Can NEET MCQs trick you with Okazaki fragment direction (3’→5’ vs 5’→3’)?
Yes, NEET questions can test understanding of DNA synthesis direction. Remember, DNA polymerase synthesizes DNA strictly in the 5’ to 3’ direction. Okazaki fragments reflect discontinuous synthesis on a template strand running 5’ to 3’, but the fragments themselves elongate 5’ to 3’. Careful reading is essential to avoid mistakes.
8. How to avoid mixing up leading and lagging strands in diagrams?
To avoid confusion, first identify the direction of the replication fork and the template strand orientations. The leading strand runs 3’ to 5’ on the template and synthesizes continuously toward the fork; the lagging strand runs 5’ to 3’ on the template and forms Okazaki fragments discontinuously away from the fork. Using arrows and labels aids clarity.
9. Are Okazaki fragments ever formed on the leading strand?
No, Okazaki fragments are formed exclusively on the lagging strand. The leading strand is synthesized continuously in the 5’ to 3’ direction as the replication fork progresses, so it does not require fragmentary synthesis.
10. How to quickly check if a question is about Okazaki fragments or primers?
To differentiate, focus on context: RNA primers initiate DNA synthesis for each Okazaki fragment; questions about initiation or primer removal relate to primers, while questions about discontinuous DNA segments joined after primer removal relate to Okazaki fragments. Keywords like “joining fragments” hint toward Okazaki fragments.
11. What is the role of RNA primers in Okazaki fragment synthesis?
RNA primers serve as starting points for DNA synthesis on the lagging strand. Since DNA polymerase cannot initiate synthesis de novo, primase lays down short RNA primers on which DNA polymerase extends the Okazaki fragments. Later, primers are removed and replaced by DNA.
12. Who discovered Okazaki fragments?
Okazaki fragments were discovered by Japanese molecular biologists Reiji and Tsuneko Okazaki in the 1960s. Their pulse-labeling experiments with radioactive thymidine in E. coli revealed short DNA segments synthesized discontinuously during replication, leading to the concept of lagging strand synthesis.