
Write the types of restriction enzymes. “Synthesis of recombinant DNA molecules is possible only when the vector and source DNA is cut by the same restriction enzyme” explain the reason.
Answer
569.1k+ views
Hint: A restriction enzyme is proteins that recognise a specific and short nucleotide sequence and cut the DNA only at the specific site, which is called the target sequence or restriction site.
Complete answer:
The restriction endonuclease is an enzyme that can recognize the specific bases sequence in the DNA molecules and cut the DNA at that site. They are also called a restriction enzyme. Depending upon the cofactor enzyme requirement, structure and composition, cleavage site, and nature of the target DNA, these are categorised into four types that are type I, type II, type III and type IV.
Type I: Cleave the target DNA sequence at random length (approximately 1000bp away) from the recognition site. It requires the cofactor S-AdenosylMethionine, ATP, and \[{\text{M}}{{\text{g}}^ + }\] to perform their activities.
Type II: They cleave mostly within or near a short distance from a recognition site. They need ${\text{M}}{{\text{g}}^ + }$ as a cofactor. The recognition site is 4-8 nucleotides in length and also undivided and palindromic.
Type III: They cut the DNA after 20-30 base pairs at the recognition site. They contain more than one subunit. To perform they required ATP and AdoMet as cofactors for DNA restriction and methylation.
Type IV: Only cleave or cut the modified DNA. Cleavage takes place approximately 30 bp away from the site.
Restriction enzymes are very useful in genetic engineering, and help to create the recombinant DNA. To create the recombinant DNA, the target DNA sequence needs to be recognised and cut. This is done with the help of restriction endonucleases enzymes. When the enzyme cut the DNA sequence it created the “sticky end”. Further, this DNA fragment will join to the vector DNA, therefore, to join the DNA fragment with the vector; the vector is also cut with the same restriction enzyme so that it creates the same “sticky end” and makes it easier to join the ends.
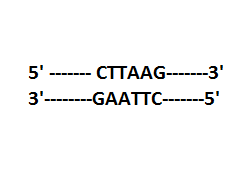
Note: Each restriction enzyme recognises a specific palindromic sequence in the DNA. The palindromes in DNA are a sequence of base pairs that read the same on the two strands that are the same in $5' \to 3'$ direction and $3' \to 5'$ direction.
Complete answer:
The restriction endonuclease is an enzyme that can recognize the specific bases sequence in the DNA molecules and cut the DNA at that site. They are also called a restriction enzyme. Depending upon the cofactor enzyme requirement, structure and composition, cleavage site, and nature of the target DNA, these are categorised into four types that are type I, type II, type III and type IV.
Type I: Cleave the target DNA sequence at random length (approximately 1000bp away) from the recognition site. It requires the cofactor S-AdenosylMethionine, ATP, and \[{\text{M}}{{\text{g}}^ + }\] to perform their activities.
Type II: They cleave mostly within or near a short distance from a recognition site. They need ${\text{M}}{{\text{g}}^ + }$ as a cofactor. The recognition site is 4-8 nucleotides in length and also undivided and palindromic.
Type III: They cut the DNA after 20-30 base pairs at the recognition site. They contain more than one subunit. To perform they required ATP and AdoMet as cofactors for DNA restriction and methylation.
Type IV: Only cleave or cut the modified DNA. Cleavage takes place approximately 30 bp away from the site.
Restriction enzymes are very useful in genetic engineering, and help to create the recombinant DNA. To create the recombinant DNA, the target DNA sequence needs to be recognised and cut. This is done with the help of restriction endonucleases enzymes. When the enzyme cut the DNA sequence it created the “sticky end”. Further, this DNA fragment will join to the vector DNA, therefore, to join the DNA fragment with the vector; the vector is also cut with the same restriction enzyme so that it creates the same “sticky end” and makes it easier to join the ends.
Note: Each restriction enzyme recognises a specific palindromic sequence in the DNA. The palindromes in DNA are a sequence of base pairs that read the same on the two strands that are the same in $5' \to 3'$ direction and $3' \to 5'$ direction.

Recently Updated Pages
Master Class 12 Economics: Engaging Questions & Answers for Success

Master Class 12 Physics: Engaging Questions & Answers for Success

Master Class 12 English: Engaging Questions & Answers for Success

Master Class 12 Social Science: Engaging Questions & Answers for Success

Master Class 12 Maths: Engaging Questions & Answers for Success

Master Class 12 Business Studies: Engaging Questions & Answers for Success

Trending doubts
Which are the Top 10 Largest Countries of the World?

What are the major means of transport Explain each class 12 social science CBSE

Draw a labelled sketch of the human eye class 12 physics CBSE

Why cannot DNA pass through cell membranes class 12 biology CBSE

Differentiate between insitu conservation and exsitu class 12 biology CBSE

Draw a neat and well labeled diagram of TS of ovary class 12 biology CBSE




